Physics-based machine learning for stochastic reaction networks
Methods and papers related to physics-based maching learning for modeling stochastic reaction networks, as developed in the thesis of Oliver K. Ernst
Research groups:
Eric Mjolsness Departments of Computer Science and Mathematics, UC Irvine
GitHub:
Click here to browse all code through this organization's GitHub page.
Physics-based machine learning for modeling stochastic IP3-dependent calcium dynamics
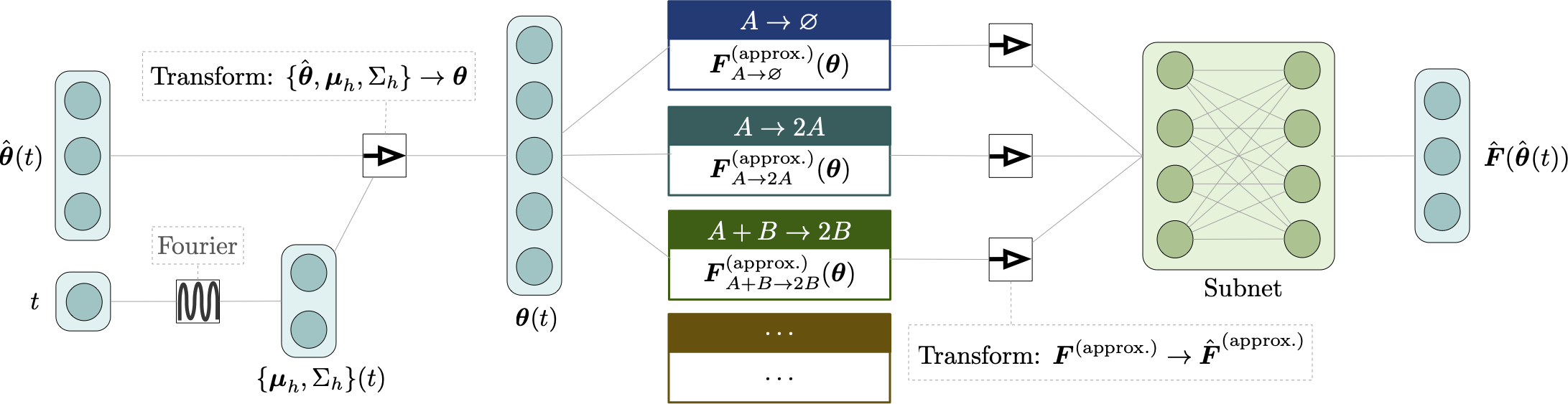
Abstract
We present a machine learning method for model reduction which incorporates domain-specific physics through candidate functions. Our method estimates an effective probability distribution and differential equation model from stochastic simulations of a reaction network. The close connection between reduced and fine scale descriptions allows approximations derived from the master equation to be introduced into the learning problem. This representation is shown to improve generalization and allows a large reduction in network size for a classic model of inositol trisphosphate (IP3) dependent calcium oscillations in non-excitable cells.
Code & directions
A complete description of the code can be found on the GitHub page’s Readme files.
Requirements
- Requires Mathematica v12 or later
- Data version control (DVC) to download data generated for the paper (else, you will have to run everything from scratch!):
brew install dvc
Download
Either:
- Grab the latest release .zip, or
- Clone the repo:
git clone https://github.com/smrfeld/physics-based-ml-reaction-networks/releases

